
A Data Wrangling in R
In this part of our toolkit, we’re going to learn how to do the same things we did with Chapter 4 - Spatial Data Wrangling, but this time, we’ll use R code to handle our spatial data.
Getting Started
R is a great choice for starting in data science because it’s built for it. It’s not just a programming language, it is a whole system with tools and libraries made to help you think and work like a data scientist easily.
We assume a basic knowledge of R and coding languages for these toolkits. For most of the tutorials in this toolkit, you’ll need to have R and RStudio downloaded and installed on your system. You should be able to install packages, know how to find the address to a folder on your computer system, and have very basic familiarity with R.
Tutorials for R
If you are new to R, we recommend the following intro-level tutorials provided through installation guides. You can also refer to this R for Social Scientists tutorial developed by Data Carpentry for a refresher.
You can also visit the RStudio Education page to select a learning path tailored to your experience level (Beginners, Intermediates, Experts). They offer detailed instructions to learners at different stages of their R journey.
A.1 Environment Setup
Getting started with data analysis in R involves a few preliminary steps, including downloading datasets and setting up a working directory. This introduction will guide you through these essential steps to ensure a smooth start to your data analysis journey in R.
Download the Activity Datasets
Please download and unzip this file to get started: SDOHPlace-DataWrangling.zip
Setting Up the Working Directory
Setting up a working directory in R is crucial–any file paths you use in your code, like the names of files to read data from, will be treated at relative to your current working directly. You should set the working directory to any folder on your system where you plan to store your datasets and R scripts.
To set your working directory, use
To see what your working directory is currently set to, use getwd().
A.1.0.1 Example: Local directory on Windows
You can use forward slashes for directory paths:
or you can use double backslashes:
A.1.0.2 Example: Local directory on macOS or Linux
Use forward slashes:
You can also use ~ as an abbreviation for your own home directory:
A.1.0.3 Example: Posit Cloud
Posit Cloud is a web environment that allows you to create R projects online. In a Posit project, your working directory will already be set to your project. You can confirm this like so:
Installing & Working with R Libraries
Before starting operations related to spatial data, we need to complete an environmental setup. This workshop requires several packages, which can be installed from CRAN:
sf: simplifies spatial data manipulationtmap: streamlines thematic map creationdplyr: facilitates data manipulationtidygeocoder: converts addresses to coordinates easily
You only need to install packages once in an R environment. Use these commands to install the packages needed for this tutorial.
install.packages("sf")
install.packages("tmap")
install.packages("tidygeocoder")
install.packages("dplyr")Installation Tip
For Mac users, check out https://github.com/r-spatial/sf for additional tips if you run into errors when installing the sf package. Using homebrew to install gdal usually fixes any remaining issues.
Now, loading the required libraries for further steps:
A.2 Intro to Spatial Data
Spatial data analysis in R provides a robust framework for understanding geographical information, enabling users to explore, visualize, and model spatial relationships directly within their data. Through the integration of specialized packages like sf for spatial data manipulation, ggplot2 and tmap for advanced mapping, and tidygeocoder for geocoding, R becomes a powerful tool for geographic data science. This ecosystem allows researchers and analysts to uncover spatial patterns, analyze geographic trends, and produce detailed maps that convey complex information intuitively.
Load Spatial Data
We need to load the spatial data (shapefile). Remember, this type of data is actually comprised of multiple files. All need to be present in order to read correctly. Let’s use chicagotracts.shp for practice, which includes the census tracts boundary in Chicago.
First, we need to read the shapefile data from where you save it.
Your output will look something like:
## Reading layer `chicagotracts' from data source `./SDOHPlace-DataWrangling/chicagotracts.shp' using driver `ESRI Shapefile'
## Simple feature collection with 801 features and 9 fields
## Geometry type: POLYGON
## Dimension: XY
## Bounding box: xmin: -87.94025 ymin: 41.64429 xmax: -87.52366 ymax: 42.02392
## Geodetic CRS: WGS 84Always inspect data when loading in. Let’s look at a non-spatial view.
## Simple feature collection with 6 features and 9 fields
## Geometry type: POLYGON
## Dimension: XY
## Bounding box: xmin: -87.68822 ymin: 41.72902 xmax: -87.62394 ymax: 41.87455
## Geodetic CRS: WGS 84
## commarea commarea_n countyfp10 geoid10 name10 namelsad10 notes statefp10 tractce10 geometry
## 1 44 44 031 17031842400 8424 Census Tract 8424 <NA> 17 842400 POLYGON ((-87.62405 41.7302...
## 2 59 59 031 17031840300 8403 Census Tract 8403 <NA> 17 840300 POLYGON ((-87.68608 41.8229...
## 3 34 34 031 17031841100 8411 Census Tract 8411 <NA> 17 841100 POLYGON ((-87.62935 41.8528...
## 4 31 31 031 17031841200 8412 Census Tract 8412 <NA> 17 841200 POLYGON ((-87.68813 41.8556...
## 5 32 32 031 17031839000 8390 Census Tract 8390 <NA> 17 839000 POLYGON ((-87.63312 41.8744...
## 6 28 28 031 17031838200 8382 Census Tract 8382 <NA> 17 838200 POLYGON ((-87.66782 41.8741...Check out the data structure of this file.
## Classes 'sf' and 'data.frame': 801 obs. of 10 variables:
## $ commarea : chr "44" "59" "34" "31" ...
## $ commarea_n: num 44 59 34 31 32 28 65 53 76 77 ...
## $ countyfp10: chr "031" "031" "031" "031" ...
## $ geoid10 : chr "17031842400" "17031840300" "17031841100" "17031841200" ...
## $ name10 : chr "8424" "8403" "8411" "8412" ...
## $ namelsad10: chr "Census Tract 8424" "Census Tract 8403" "Census Tract 8411" "Census Tract 8412" ...
## $ notes : chr NA NA NA NA ...
## $ statefp10 : chr "17" "17" "17" "17" ...
## $ tractce10 : chr "842400" "840300" "841100" "841200" ...
## $ geometry :sfc_POLYGON of length 801; first list element: List of 1
## ..$ : num [1:243, 1:2] -87.6 -87.6 -87.6 -87.6 -87.6 ...
## ..- attr(*, "class")= chr [1:3] "XY" "POLYGON" "sfg"
## - attr(*, "sf_column")= chr "geometry"
## - attr(*, "agr")= Factor w/ 3 levels "constant","aggregate",..: NA NA NA NA NA NA NA NA NA
## ..- attr(*, "names")= chr [1:9] "commarea" "commarea_n" "countyfp10" "geoid10" ...The data is no longer a shapefile but an sf object, comprised of polygons. The plot() command in R help to quickly visualizes the geometric shapes of Chicago’s census tracts. The output includes multiple maps because the sf framework enables previews of each attribute in our spatial file.

A.2.1 Adding a Basemap
Then, we can use tmap, a mapping library, in interactive mode to add a basemap layer. It plots the spatial data from Chi_tracts, applies a minimal theme for clarity, and labels the map with a title, offering a straightforward visualization of the area’s census tracts.
We stylize the borders of the tract boundaries by making it transparent at 50% (which is equal to an alpha level of 0.5).
## ℹ tmap mode set to "view".##
## ── tmap v3 code detected ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
## [v3->v4] `tm_borders()`: use `fill_alpha` instead of `alpha`.[v3->v4] `tm_layout()`: use `tm_title()` instead of `tm_layout(title = )`Still in the interactive mode (`view’), we can switch to a different basemap. Here we bring in a “Voyager” style map from Carto, a cartographic firm. We’ll make the borders less transparent by adjusting the alpha level.
## ℹ tmap mode set to "view".tm_basemap("CartoDB.Voyager") +
tm_shape(Chi_tracts) + tm_borders(alpha=0.8, col = "gray40") +
tm_layout(title = "Census Tract Map of Chicago")##
## ── tmap v3 code detected ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
## [v3->v4] `tm_borders()`: use `fill_alpha` instead of `alpha`.[v3->v4] `tm_layout()`: use `tm_title()` instead of `tm_layout(title = )`Tip
For additional options, you can preview basemaps at the Leaflet Providers Demo. Some basemaps we recommend that work consistently are by:
- CartoDB (Carto)
- Open Street Map
- ESRI
Not all basemaps are available anymore, and some require keys that you’d need to add on your own.
A.3 Coordinate Reference Systems
For this exercise we will use chicagotracts.shp to explore how to change the projection of a spatial dataset in R. First, let’s check out the current coordinate reference system.
## Coordinate Reference System:
## User input: WGS 84
## wkt:
## GEOGCRS["WGS 84",
## DATUM["World Geodetic System 1984",
## ELLIPSOID["WGS 84",6378137,298.257223563,
## LENGTHUNIT["metre",1]]],
## PRIMEM["Greenwich",0,
## ANGLEUNIT["degree",0.0174532925199433]],
## CS[ellipsoidal,2],
## AXIS["latitude",north,
## ORDER[1],
## ANGLEUNIT["degree",0.0174532925199433]],
## AXIS["longitude",east,
## ORDER[2],
## ANGLEUNIT["degree",0.0174532925199433]],
## ID["EPSG",4326]]We can use the st_transform function to transform CRS. When projecting a dataset of Illinois, the most appropriate NAD83 projection would be NAD83 UTM zone 16N. Chicago sits within the area best covered by NAD83 / Illinois East (ftUS) (EPSG:3435).
Chi_tracts.3435 <- st_transform(Chi_tracts, "EPSG:3435")
# Chi_tracts.3435 <- st_transform(Chi_tracts, 3435)
st_crs(Chi_tracts.3435)## Coordinate Reference System:
## User input: EPSG:3435
## wkt:
## PROJCRS["NAD83 / Illinois East (ftUS)",
## BASEGEOGCRS["NAD83",
## DATUM["North American Datum 1983",
## ELLIPSOID["GRS 1980",6378137,298.257222101,
## LENGTHUNIT["metre",1]]],
## PRIMEM["Greenwich",0,
## ANGLEUNIT["degree",0.0174532925199433]],
## ID["EPSG",4269]],
## CONVERSION["SPCS83 Illinois East zone (US survey foot)",
## METHOD["Transverse Mercator",
## ID["EPSG",9807]],
## PARAMETER["Latitude of natural origin",36.6666666666667,
## ANGLEUNIT["degree",0.0174532925199433],
## ID["EPSG",8801]],
## PARAMETER["Longitude of natural origin",-88.3333333333333,
## ANGLEUNIT["degree",0.0174532925199433],
## ID["EPSG",8802]],
## PARAMETER["Scale factor at natural origin",0.999975,
## SCALEUNIT["unity",1],
## ID["EPSG",8805]],
## PARAMETER["False easting",984250,
## LENGTHUNIT["US survey foot",0.304800609601219],
## ID["EPSG",8806]],
## PARAMETER["False northing",0,
## LENGTHUNIT["US survey foot",0.304800609601219],
## ID["EPSG",8807]]],
## CS[Cartesian,2],
## AXIS["easting (X)",east,
## ORDER[1],
## LENGTHUNIT["US survey foot",0.304800609601219]],
## AXIS["northing (Y)",north,
## ORDER[2],
## LENGTHUNIT["US survey foot",0.304800609601219]],
## USAGE[
## SCOPE["Engineering survey, topographic mapping."],
## AREA["United States (USA) - Illinois - counties of Boone; Champaign; Clark; Clay; Coles; Cook; Crawford; Cumberland; De Kalb; De Witt; Douglas; Du Page; Edgar; Edwards; Effingham; Fayette; Ford; Franklin; Gallatin; Grundy; Hamilton; Hardin; Iroquois; Jasper; Jefferson; Johnson; Kane; Kankakee; Kendall; La Salle; Lake; Lawrence; Livingston; Macon; Marion; Massac; McHenry; McLean; Moultrie; Piatt; Pope; Richland; Saline; Shelby; Vermilion; Wabash; Wayne; White; Will; Williamson."],
## BBOX[37.06,-89.27,42.5,-87.02]],
## ID["EPSG",3435]]After change the projection, we can plot the map. We’ll switch to the static version of tmap, using the plot mode.
## ℹ tmap mode set to "plot".tm_shape(Chi_tracts.3435) + tm_borders(alpha=0.5) +
tm_layout(main.title ="EPSG:3435 (ft)",
main.title.position = "center")##
## ── tmap v3 code detected ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
## [v3->v4] `tm_borders()`: use `fill_alpha` instead of `alpha`.[v3->v4] `tm_layout()`: use `tm_title()` instead of `tm_layout(main.title = )`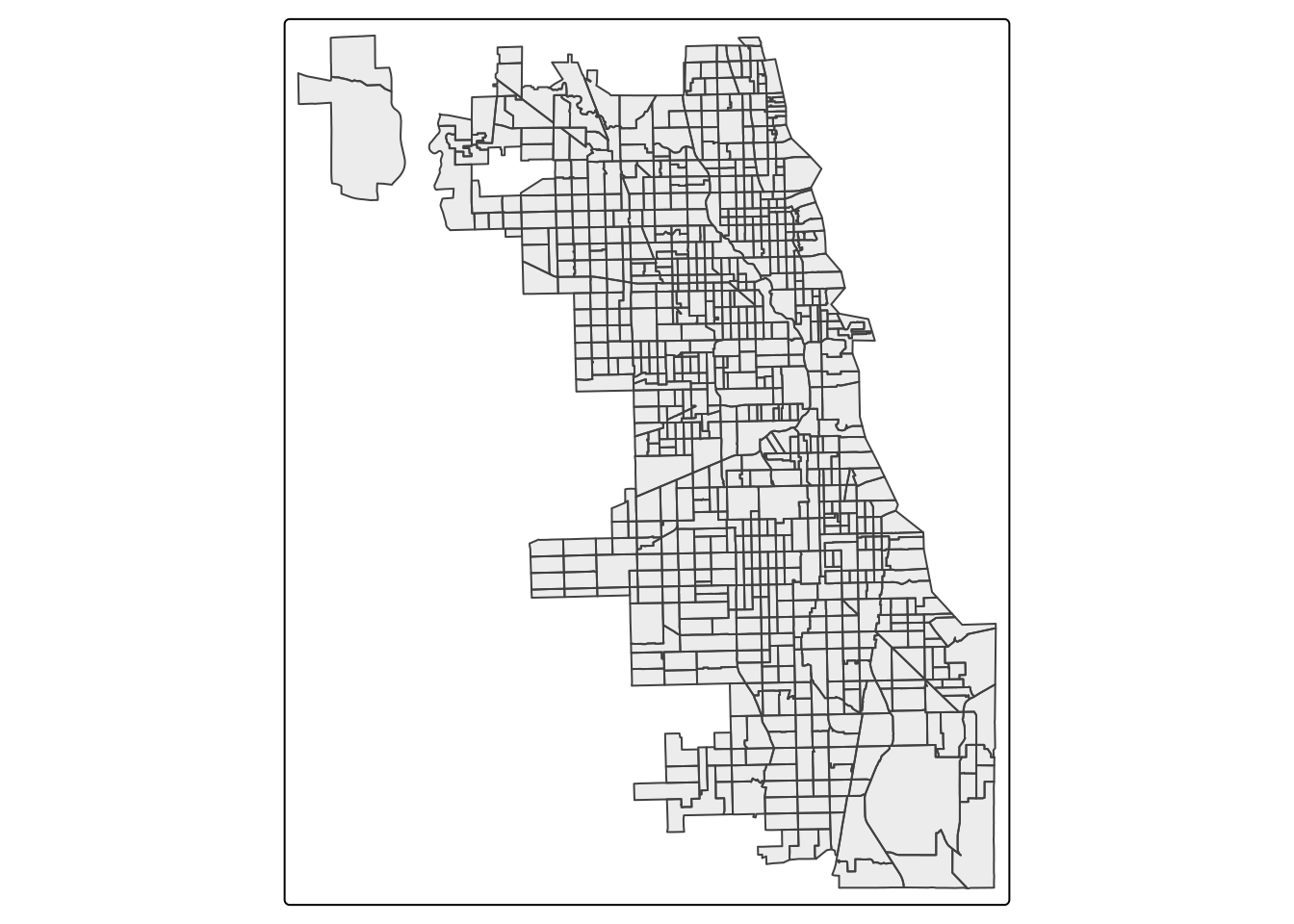
What if we had used the wrong EPSG code, referencing the wrong projection? Here we’ll transform and plot EPSG Code 3561, a coordinate reference system in Hawai’i.
Chi_tracts.3561 <- st_transform(Chi_tracts, "EPSG:3561")
tm_shape(Chi_tracts.3561) + tm_borders(alpha=0.5) +
tm_layout(main.title ="EPSG:3561 (ft)",
main.title.position = "center")## ## ── tmap v3 code detected ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────## [v3->v4] `tm_borders()`: use `fill_alpha` instead of `alpha`.
## [v3->v4] `tm_layout()`: use `tm_title()` instead of `tm_layout(main.title = )`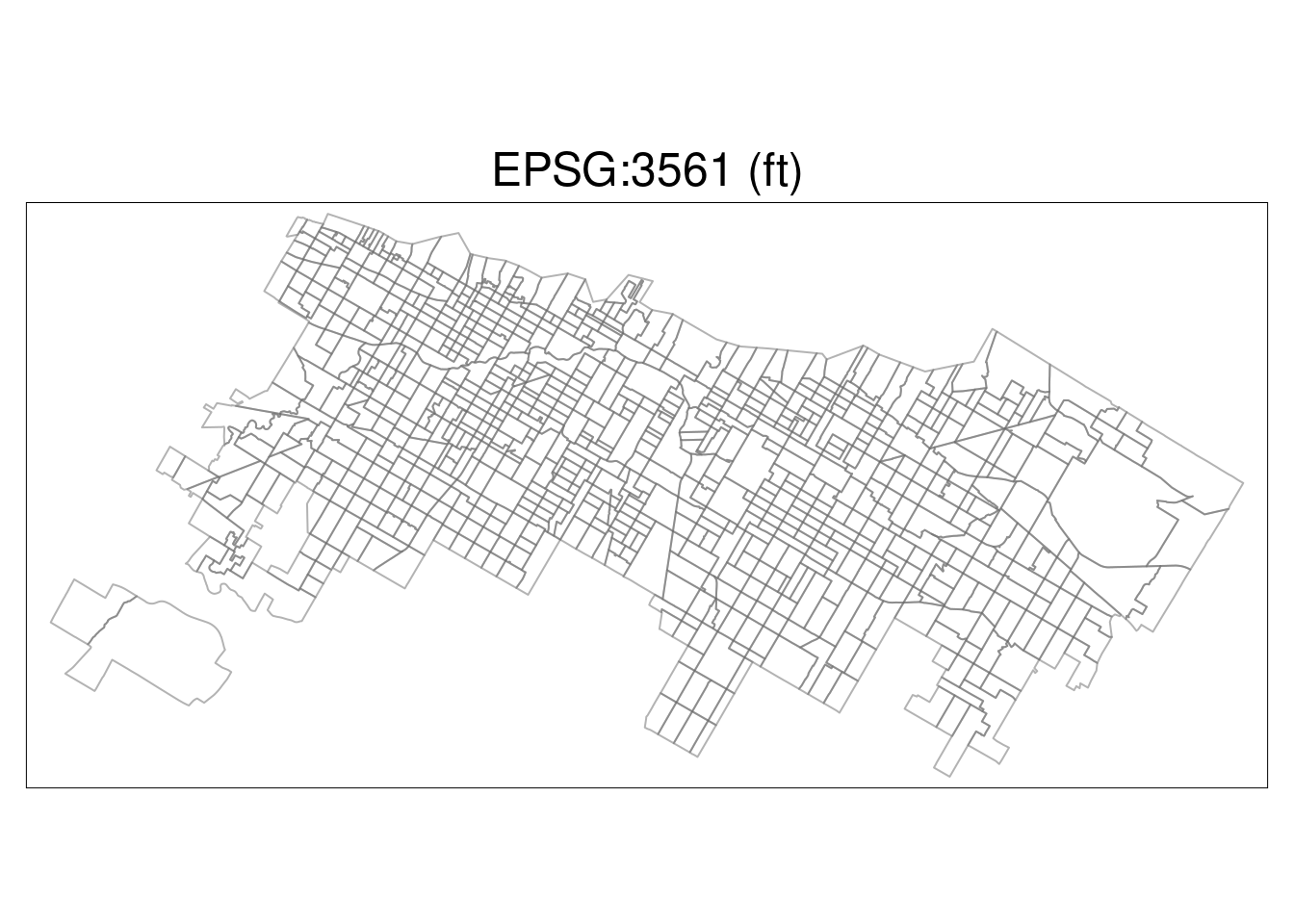
It’s obviously not correct – the wrong CRS can cause a load of trouble. Make sure you specify carefully!
Refine Basic Map
Let’s take a deeper look at the cartographic mapping package, tmap. We approach mapping with one layer at a time. Always start with the object you want to map by calling it with the tm_shape function. Then, at least one descriptive/styling function follows. There are hundreds of variations and paramater specification.
Here we style the tracts with some semi-transparent borders.
## ## ── tmap v3 code detected ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────## [v3->v4] `tm_borders()`: use `fill_alpha` instead of `alpha`.
Next we fill the tracts with a light gray, and adjust the color and transparency of borders. We also add a scale bar, positioning it to the left and having a thickness of 0.8 units, and turn off the frame.
tm_shape(Chi_tracts) + tm_fill(col = "gray90") + tm_borders(alpha=0.2, col = "gray10") +
tm_scale_bar(position = ("bottom"), lwd = 0.8) +
tm_layout(frame = F)## ## ── tmap v3 code detected ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────## [v3->v4] `tm_borders()`: use `fill_alpha` instead of `alpha`.
## ! `tm_scale_bar()` is deprecated. Please use `tm_scalebar()` instead.
Check out https://rdrr.io/cran/tmap/man/tm_polygons.html for more ideas.
A.4 Converting to Spatial Data
A.4.1 Convert CSVs to Spatial Data
We are using the Affordable_Rental_Housing_Developments.csv in the dataset to show how to convert a csv Lat/Long data to points. First, we need to load the CSV data.
Then, we need to ensure that no column (intended to be used as a coordinate) is entirely empty or filled with NA values.
Inspect the data to confirm it’s doing what you expect it to be doing. What columns will you use to specify the coordinates? In this dataset, we have multiple coordinate options. We’ll use latitude and longitude, or rather, longitude as our X value, and latitude as our Y value. In the data, it’s specified as “Longitude” and “Latitude.”
## Community.Area.Name Community.Area.Number Property.Type Property.Name Address Zip.Code Phone.Number Management.Company Units
## 2 Rogers Park 1 Senior Morse Senior Apts. 6928 N. Wayne Ave. 60626 312-602-6207 Morse Urban Dev. 44
## 3 Uptown 3 ARO The Draper 5050 N. Broadway 60640 312-818-1722 Flats LLC 35
## 4 Edgewater 77 Senior Pomeroy Apts. 5650 N. Kenmore Ave. 60660 773-275-7820 Habitat Company 198
## 5 Roseland 49 Supportive Housing Wentworth Commons 11045 S. Wentworth Ave. 60628 773-568-7804 Mercy Housing Lakefront 50
## 6 Humboldt Park 23 Multifamily Nelson Mandela Apts. 607 N. Sawyer Ave. 60624 773-227-6332 Bickerdike Apts. 6
## 7 Grand Boulevard 38 Multifamily Legends South - Gwendolyn Place 4333 S. Michigan Ave. 60653 773-624-7676 Interstate Realty Management Co. 71
## X.Coordinate Y.Coordinate Latitude Longitude Location
## 2 1165844 1946059 42.00757 -87.66517 (42.0075737709331, -87.6651711448293)
## 3 1167357 1933882 41.97413 -87.65996 (41.9741295261027, -87.6599553011627)
## 4 1168181 1937918 41.98519 -87.65681 (41.9851867755403, -87.656808676983)
## 5 1176951 1831516 41.69302 -87.62777 (41.6930159120977, -87.6277673462214)
## 6 1154640 1903912 41.89215 -87.70753 (41.8921534052465, -87.7075265659001)
## 7 1177924 1876178 41.81555 -87.62286 (41.815550396096, -87.6228565224104)Finally, we start to convert it to points. Be sure you use the CRS of the original coordinates recorded. In this case we weren’t sure what CRS that was, so we use EPSG:4326 to test.
View the resulting sf object with a basemap to confirm they are in the right place. Overlay them on top of the tract data, to confirm they are plotting correctly.
### First Layer
tm_shape(Chi_tracts) + tm_borders(lwd = 0.5) +
### Second Layer
tm_shape(points_housing) + tm_dots(size = 0.1 )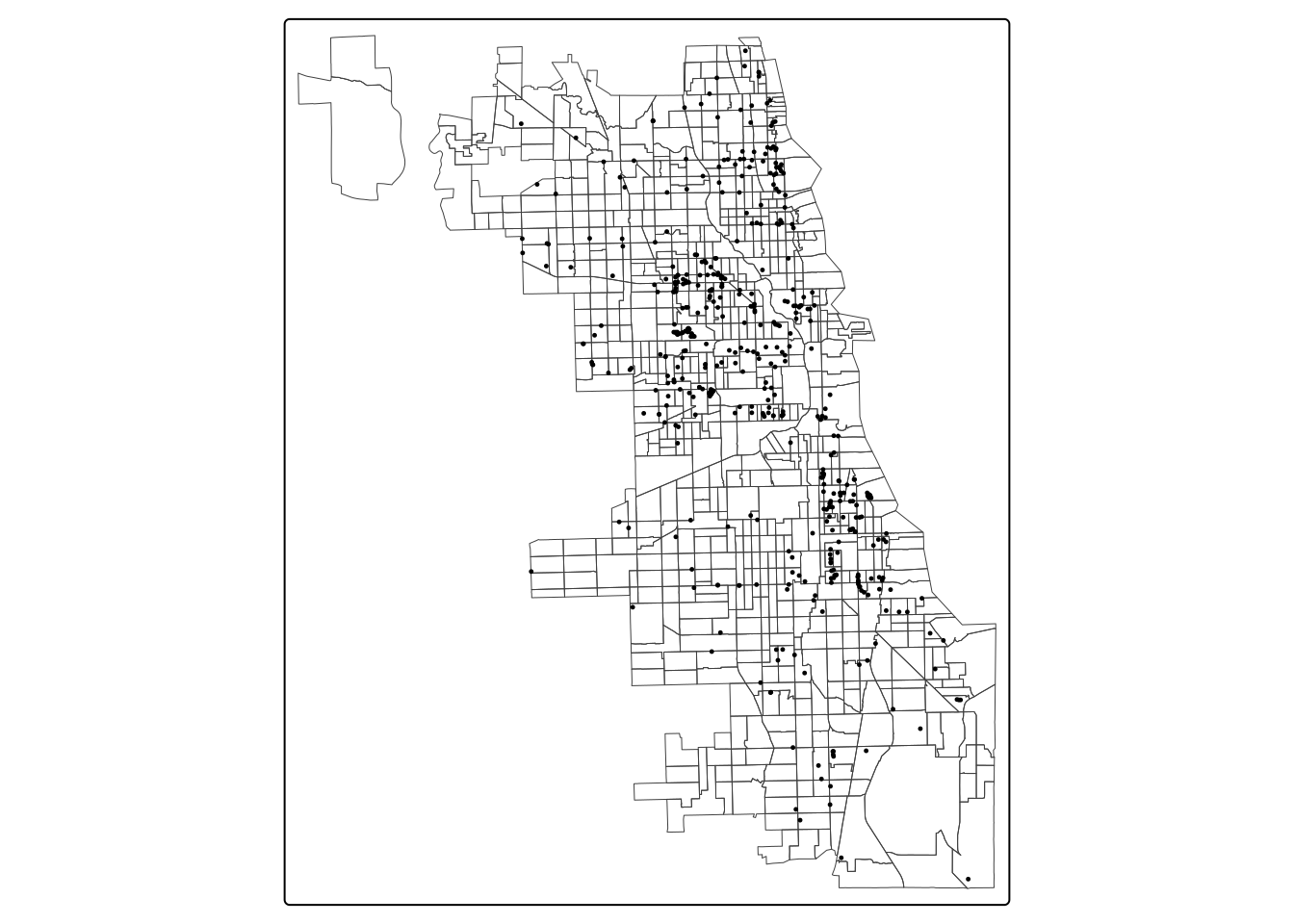
You can change the tmap_mode to “view” to add a basemap in an interactive setting, and then switch back to “plot” when complete. Because we’e plotting dots using tmap, we’ll use the tm_dots parameter for styling.
## ℹ tmap mode set to "view".## ℹ tmap mode set to "plot".We’ll reproject to EPSG:3435, our system standard
A.4.1.1 Write Spatial Data
Finally, we can save our points as a spatial dataset. Use ‘st_write’ to write your spatial object in R to a data format of your choice. Here, we’ll write to a geojson file.
You could also save as a “housing.shapefile” to get a shapefile format, however you’ll get an error noting that some column names are too long and must be shortened. Shapefiles have a limit of 10 characters for field names.
The file may still write, but the column names that were too long may be shortened automatically.
To change column or field names in R objects, there are dozens of options. Try searching and “Googling” different search terms to identify solutions on your own.
A.4.2 Geocode Addresses
Here, we will use chicago_methadone_nogeometry.csv for practice, which includes methadone centers in Chicago (center names and addresses). First we load the tidygeocoder library to get our geocoding done.
Let’s read in and inspect data for methadone maintenance providers. Note, these addresses were made available by SAMHSA, and are known as publicly available information. An additional analysis could call each service to check on access to medication during COVID in September 2020, and the list would be updated further.
methadoneClinics <- read.csv("SDOHPlace-DataWrangling/chicago_methadone_nogeometry.csv")
head(methadoneClinics)## X Name Address City State Zip
## 1 1 Chicago Treatment and Counseling Center, Inc. 4453 North Broadway st. Chicago IL 60640
## 2 2 Sundace Methadone Treatment Center, LLC 4545 North Broadway St. Chicago IL 60640
## 3 3 Soft Landing Interventions/DBA Symetria Recovery of Lakeview 3934 N. Lincoln Ave. Chicago IL 60613
## 4 4 PDSSC - Chicago, Inc. 2260 N. Elston Ave. Chicago IL 60614
## 5 5 Center for Addictive Problems, Inc. 609 N. Wells St. Chicago IL 60654
## 6 6 Family Guidance Centers, Inc. 310 W. Chicago Ave. Chicago IL 60654Let’s geocode one address first, just to make sure our system is working. We’ll use the “osm” method to use the OpenStreetMap geocoder.
## Passing 1 address to the Nominatim single address geocoder## Query completed in: 1 seconds## # A tibble: 1 × 3
## address latitude longitude
## <chr> <dbl> <dbl>
## 1 2260 N. Elston Ave. Chicago, IL 41.9 -87.7As we prepare for geocoding, check out the structure of the dataset. The data should be a character to be read properly.
## 'data.frame': 27 obs. of 6 variables:
## $ X : int 1 2 3 4 5 6 7 8 9 10 ...
## $ Name : chr "Chicago Treatment and Counseling Center, Inc." "Sundace Methadone Treatment Center, LLC" "Soft Landing Interventions/DBA Symetria Recovery of Lakeview" "PDSSC - Chicago, Inc." ...
## $ Address: chr "4453 North Broadway st." "4545 North Broadway St." "3934 N. Lincoln Ave." "2260 N. Elston Ave." ...
## $ City : chr "Chicago" "Chicago" "Chicago" "Chicago" ...
## $ State : chr "IL" "IL" "IL" "IL" ...
## $ Zip : int 60640 60640 60613 60614 60654 60654 60651 60607 60607 60616 ...We need to clean the data a bit. We’ll add a new column for a full address, as required by the geocoding service. When you use a geocoding service, be sure to read the documentation and understand how the data needs to be formatted for input.
methadoneClinics$fullAdd <- paste(as.character(methadoneClinics$Address),
as.character(methadoneClinics$City),
as.character(methadoneClinics$State),
as.character(methadoneClinics$Zip))We’re ready to go! Batch geocode with one function, and inspect:
geoCodedClinics <- geocode(methadoneClinics,
address = 'fullAdd', lat = latitude, long = longitude, method="osm")## Passing 27 addresses to the Nominatim single address geocoder## Query completed in: 37.3 seconds## # A tibble: 6 × 9
## X Name Address City State Zip fullAdd latitude longitude
## <int> <chr> <chr> <chr> <chr> <int> <chr> <dbl> <dbl>
## 1 1 Chicago Treatment and Counseling Center, Inc. 4453 North Broadway st. Chicago IL 60640 4453 North Broadway st. Chicago IL 60640 42.0 -87.7
## 2 2 Sundace Methadone Treatment Center, LLC 4545 North Broadway St. Chicago IL 60640 4545 North Broadway St. Chicago IL 60640 42.0 -87.7
## 3 3 Soft Landing Interventions/DBA Symetria Recovery of Lakeview 3934 N. Lincoln Ave. Chicago IL 60613 3934 N. Lincoln Ave. Chicago IL 60613 42.0 -87.7
## 4 4 PDSSC - Chicago, Inc. 2260 N. Elston Ave. Chicago IL 60614 2260 N. Elston Ave. Chicago IL 60614 41.9 -87.7
## 5 5 Center for Addictive Problems, Inc. 609 N. Wells St. Chicago IL 60654 609 N. Wells St. Chicago IL 60654 41.9 -87.6
## 6 6 Family Guidance Centers, Inc. 310 W. Chicago Ave. Chicago IL 60654 310 W. Chicago Ave. Chicago IL 60654 41.9 -87.6In some cases you may find that an address does not geocode correctly. You can inspect further. This could involve a quick check for spelling issues or searching the address and pulling the lat/long using Google Maps and inputting manually. Or, if we are concerned it’s a human or unknown error, we could omit. For this exercise we’ll just omit the two clinics that didn’t geocode correctly.
A.5 Convert to Spatial Data
This is not spatial data yet! To convert a static file to spatial data, we use the powerful st_as_sf function from sf. Indicate the x,y parameters (=longitude, latitude) and the coordinate reference system used. Our geocoding service used the standard EPSG:4326, so we input that here.
Basic Map of Points
For a really simple map of points – to ensure they were geocoded and converted to spatial data correctly, we use tmap. We’ll use the interactive version to view.
If your points didn’t plot correctly:
- Did you flip the longitude/latitude values?
- Did you input the correct CRS?
Those two issues are the most common errors.
A.6 Merge Data sets
Reshape Data
Here, we are trying to use the COVID-19_Cases__Tests__and_Deaths_by_ZIP_Code.csv dataset to practice how to convert long data to a wide data format.
We subset to the first two columns, and the sixth column. That gives us the zip code, the reporting week, and cumulative cases of Covid-19. Choose whatever subset function you prefer best!
covid = read.csv("SDOHPlace-DataWrangling/COVID-19_Cases__Tests__and_Deaths_by_ZIP_Code.csv")
covid_clean = covid[,c(1:2, 6)]
head(covid_clean) ## ZIP.Code Week.Number Cases...Cumulative
## 1 60603 39 13
## 2 60604 39 31
## 3 60611 16 72
## 4 60611 15 64
## 5 60615 11 NA
## 6 60603 10 NANow, we want each zip code to be a unique row, with cases by week as a columns because we are trying to create a wide data set with the cumulative cases for each week for each zip code. Enter the code and you will see the new wide data format.
covid_wide <- reshape(covid_clean, direction = "wide",
idvar = "ZIP.Code", timevar = "Week.Number")
head(covid_wide)## ZIP.Code Cases...Cumulative.39 Cases...Cumulative.16 Cases...Cumulative.15 Cases...Cumulative.11 Cases...Cumulative.10 Cases...Cumulative.12 Cases...Cumulative.13
## 1 60603 13 NA NA NA NA NA NA
## 2 60604 31 NA NA NA NA NA NA
## 3 60611 458 72 64 NA NA 16 41
## 5 60615 644 171 132 NA NA 26 57
## 31 60605 391 93 65 NA NA 23 39
## 32 Unknown 240 23 18 NA NA NA NA
## Cases...Cumulative.14 Cases...Cumulative.34 Cases...Cumulative.17 Cases...Cumulative.18 Cases...Cumulative.19 Cases...Cumulative.20 Cases...Cumulative.31 Cases...Cumulative.22
## 1 NA 11 NA NA NA 5 9 6
## 2 NA 29 6 11 14 17 25 22
## 3 57 352 80 92 99 114 286 139
## 5 99 567 215 243 274 302 526 353
## 31 52 325 118 135 149 155 291 187
## 32 9 162 33 53 62 69 127 97
## Cases...Cumulative.23 Cases...Cumulative.24 Cases...Cumulative.25 Cases...Cumulative.28 Cases...Cumulative.29 Cases...Cumulative.30 Cases...Cumulative.32 Cases...Cumulative.33
## 1 6 6 6 6 8 9 10 11
## 2 23 24 24 25 25 25 25 25
## 3 148 152 163 223 240 264 305 333
## 5 364 376 388 444 482 506 539 556
## 31 194 198 215 247 263 277 304 312
## 32 102 106 112 120 124 126 132 142
## Cases...Cumulative.26 Cases...Cumulative.27 Cases...Cumulative.36 Cases...Cumulative.38 Cases...Cumulative.21 Cases...Cumulative.35 Cases...Cumulative.37 Cases...Cumulative.40
## 1 6 6 11 13 6 11 13 14
## 2 25 25 31 31 20 30 31 31
## 3 175 196 391 435 124 371 411 478
## 5 401 418 606 629 332 588 613 651
## 31 229 235 354 379 169 333 364 399
## 32 113 116 186 216 92 178 201 288Join by Attribute
Here, we’ll merge data sets with a common variable in R. Merging the cumulative case data set you created in the last section to zip code spatial data (ChiZipMaster1.geojson) will allow you to map the case data. You’ll be merging the case data and spatial data using the zip codes field of each dataset.
We’ve cleaned our covid case data already, but not all values under the zip code column are valid. There is a row has a value of “unknown”, so let’s remove that.
Then, we need to load the zip code data.
Your output will look something like:
Reading layer `ChiZipMaster1' from data source `./SDOHPlace-DataWrangling/ChiZipMaster1.geojson' using driver `GeoJSON'
Simple feature collection with 59 features and 30 fields
Geometry type: MULTIPOLYGON
Dimension: XY
Bounding box: xmin: -87.94011 ymin: 41.64454 xmax: -87.52414 ymax: 42.02304
Geodetic CRS: WGS 84Now, the two datasets are ready to join together by the zip code. Make sure to check they have been joined successully.
We call the these joined datasets Chi_Zipsf, to denote a final zip code master dataset.
We’ll reproject to EPSG:3435, the standard used in our study area.
Join by Location
We’ll create a spatial join with the housing and zip code data we’ve brought in.
In this example, we want to join zip-level data to the Rental Housing Developments, so we can identify which zips they are within.
First, let’s try “sticking” all the zip code data to the housing points, essentially intersecting zip codes with housing developments.
To do this, both datasets will need to be in the same CRS. We have already standardized both using EPSG:3435.
Don’t forget to inspect the data.
## Simple feature collection with 6 features and 74 fields
## Geometry type: POINT
## Dimension: XY
## Bounding box: xmin: 1165843 ymin: 1946059 xmax: 1165843 ymax: 1946059
## Projected CRS: NAD83 / Illinois East (ftUS)
## Community.Area.Name Community.Area.Number Property.Type Property.Name Address Zip.Code Phone.Number Management.Company Units X.Coordinate Y.Coordinate
## 2 Rogers Park 1 Senior Morse Senior Apts. 6928 N. Wayne Ave. 60626 312-602-6207 Morse Urban Dev. 44 1165844 1946059
## 2.1 Rogers Park 1 Senior Morse Senior Apts. 6928 N. Wayne Ave. 60626 312-602-6207 Morse Urban Dev. 44 1165844 1946059
## 2.2 Rogers Park 1 Senior Morse Senior Apts. 6928 N. Wayne Ave. 60626 312-602-6207 Morse Urban Dev. 44 1165844 1946059
## 2.3 Rogers Park 1 Senior Morse Senior Apts. 6928 N. Wayne Ave. 60626 312-602-6207 Morse Urban Dev. 44 1165844 1946059
## 2.4 Rogers Park 1 Senior Morse Senior Apts. 6928 N. Wayne Ave. 60626 312-602-6207 Morse Urban Dev. 44 1165844 1946059
## 2.5 Rogers Park 1 Senior Morse Senior Apts. 6928 N. Wayne Ave. 60626 312-602-6207 Morse Urban Dev. 44 1165844 1946059
## Location zip objectid shape_area shape_len Case.Rate...Cumulative year totPopE whiteP blackP amIndP asianP pacIsP otherP hispP noHSP age0_4 age5_14
## 2 (42.0075737709331, -87.6651711448293) 60626 1 49170579 33983.91 2911.7 2018 49730 59.2 25.27 0.1 5.89 0.01 9.53 20.62 11.82 2553 4765
## 2.1 (42.0075737709331, -87.6651711448293) 60626 1 49170579 33983.91 2911.7 2018 49730 59.2 25.27 0.1 5.89 0.01 9.53 20.62 11.82 2553 4765
## 2.2 (42.0075737709331, -87.6651711448293) 60626 1 49170579 33983.91 2911.7 2018 49730 59.2 25.27 0.1 5.89 0.01 9.53 20.62 11.82 2553 4765
## 2.3 (42.0075737709331, -87.6651711448293) 60626 1 49170579 33983.91 2911.7 2018 49730 59.2 25.27 0.1 5.89 0.01 9.53 20.62 11.82 2553 4765
## 2.4 (42.0075737709331, -87.6651711448293) 60626 1 49170579 33983.91 2911.7 2018 49730 59.2 25.27 0.1 5.89 0.01 9.53 20.62 11.82 2553 4765
## 2.5 (42.0075737709331, -87.6651711448293) 60626 1 49170579 33983.91 2911.7 2018 49730 59.2 25.27 0.1 5.89 0.01 9.53 20.62 11.82 2553 4765
## age15_19 age20_24 age15_44 age45_49 age50_54 age55_59 age60_64 ageOv65 ageOv18 age18_64 a15_24P und45P ovr65P disbP Cases...Cumulative.39 Cases...Cumulative.16
## 2 2937 5056 25875 3295 3051 2713 2447 5031 41428 36397 16.07 66.75 10.12 10.8 1448 301
## 2.1 2937 5056 25875 3295 3051 2713 2447 5031 41428 36397 16.07 66.75 10.12 10.8 1448 301
## 2.2 2937 5056 25875 3295 3051 2713 2447 5031 41428 36397 16.07 66.75 10.12 10.8 1448 301
## 2.3 2937 5056 25875 3295 3051 2713 2447 5031 41428 36397 16.07 66.75 10.12 10.8 1448 301
## 2.4 2937 5056 25875 3295 3051 2713 2447 5031 41428 36397 16.07 66.75 10.12 10.8 1448 301
## 2.5 2937 5056 25875 3295 3051 2713 2447 5031 41428 36397 16.07 66.75 10.12 10.8 1448 301
## Cases...Cumulative.15 Cases...Cumulative.11 Cases...Cumulative.10 Cases...Cumulative.12 Cases...Cumulative.13 Cases...Cumulative.14 Cases...Cumulative.34 Cases...Cumulative.17
## 2 166 NA NA 17 40 83 1352 501
## 2.1 166 NA NA 17 40 83 1352 501
## 2.2 166 NA NA 17 40 83 1352 501
## 2.3 166 NA NA 17 40 83 1352 501
## 2.4 166 NA NA 17 40 83 1352 501
## 2.5 166 NA NA 17 40 83 1352 501
## Cases...Cumulative.18 Cases...Cumulative.19 Cases...Cumulative.20 Cases...Cumulative.31 Cases...Cumulative.22 Cases...Cumulative.23 Cases...Cumulative.24 Cases...Cumulative.25
## 2 629 735 831 1279 1006 1074 1103 1118
## 2.1 629 735 831 1279 1006 1074 1103 1118
## 2.2 629 735 831 1279 1006 1074 1103 1118
## 2.3 629 735 831 1279 1006 1074 1103 1118
## 2.4 629 735 831 1279 1006 1074 1103 1118
## 2.5 629 735 831 1279 1006 1074 1103 1118
## Cases...Cumulative.28 Cases...Cumulative.29 Cases...Cumulative.30 Cases...Cumulative.32 Cases...Cumulative.33 Cases...Cumulative.26 Cases...Cumulative.27 Cases...Cumulative.36
## 2 1208 1238 1262 1296 1322 1142 1169 1384
## 2.1 1208 1238 1262 1296 1322 1142 1169 1384
## 2.2 1208 1238 1262 1296 1322 1142 1169 1384
## 2.3 1208 1238 1262 1296 1322 1142 1169 1384
## 2.4 1208 1238 1262 1296 1322 1142 1169 1384
## 2.5 1208 1238 1262 1296 1322 1142 1169 1384
## Cases...Cumulative.38 Cases...Cumulative.21 Cases...Cumulative.35 Cases...Cumulative.37 Cases...Cumulative.40 geometry
## 2 1425 913 1362 1407 1468 POINT (1165843 1946059)
## 2.1 1425 913 1362 1407 1468 POINT (1165843 1946059)
## 2.2 1425 913 1362 1407 1468 POINT (1165843 1946059)
## 2.3 1425 913 1362 1407 1468 POINT (1165843 1946059)
## 2.4 1425 913 1362 1407 1468 POINT (1165843 1946059)
## 2.5 1425 913 1362 1407 1468 POINT (1165843 1946059)We could also flip things around, and try to count how many developments intersect each zip code. We can use lengths() to find out how many items are present in a vector.
Chi_Zipsf.3435$TotHousing <- lengths(st_intersects(Chi_Zipsf.3435, housing.3435))
head(Chi_Zipsf.3435)## Simple feature collection with 6 features and 63 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: 1173037 ymin: 1889918 xmax: 1183259 ymax: 1902960
## Projected CRS: NAD83 / Illinois East (ftUS)
## zip objectid shape_area shape_len Case.Rate...Cumulative year totPopE whiteP blackP amIndP asianP pacIsP otherP hispP noHSP age0_4 age5_14 age15_19 age20_24 age15_44 age45_49
## 1 60601 27 9166246 19804.58 1451.4 2018 14675 74.17 5.57 0.45 18.00 0.00 1.81 8.68 0.00 550 156 907 909 8726 976
## 2 60602 26 4847125 14448.17 1688.1 2018 1244 68.17 3.78 5.31 19.45 0.00 3.30 6.51 0.00 61 87 18 91 987 46
## 3 60603 19 4560229 13672.68 1107.3 2018 1174 63.46 3.24 0.00 27.60 0.00 5.71 9.80 0.00 13 43 179 172 684 75
## 4 60604 48 4294902 12245.81 3964.2 2018 782 63.43 5.63 0.00 29.67 0.00 1.28 4.35 0.00 12 7 52 168 450 27
## 5 60605 20 36301276 37973.35 1420.8 2018 27519 61.20 17.18 0.18 16.10 0.03 5.31 5.84 2.39 837 1279 2172 2282 16364 1766
## 6 60606 31 6766411 12040.44 2289.6 2018 3101 72.75 2.35 0.00 18.09 0.00 6.80 6.29 0.73 57 44 0 139 1863 213
## age50_54 age55_59 age60_64 ageOv65 ageOv18 age18_64 a15_24P und45P ovr65P disbP Cases...Cumulative.39 Cases...Cumulative.16 Cases...Cumulative.15 Cases...Cumulative.11
## 1 1009 324 859 2075 13855 11780 12.37 64.27 14.14 6.4 213 36 33 NA
## 2 53 0 5 5 1095 1090 8.76 91.24 0.40 0.2 21 5 NA NA
## 3 47 150 50 112 1118 1006 29.90 63.03 9.54 7.3 13 NA NA NA
## 4 47 54 92 93 744 651 28.13 59.97 11.89 4.1 31 NA NA NA
## 5 1520 1824 1360 2569 25259 22690 16.19 67.15 9.34 5.3 391 93 65 NA
## 6 153 168 172 431 3000 2569 4.48 63.33 13.90 1.9 71 13 7 NA
## Cases...Cumulative.10 Cases...Cumulative.12 Cases...Cumulative.13 Cases...Cumulative.14 Cases...Cumulative.34 Cases...Cumulative.17 Cases...Cumulative.18 Cases...Cumulative.19
## 1 NA 14 23 28 135 43 56 63
## 2 NA NA NA NA 19 5 5 8
## 3 NA NA NA NA 11 NA NA NA
## 4 NA NA NA NA 29 6 11 14
## 5 NA 23 39 52 325 118 135 149
## 6 NA NA NA 5 58 15 17 19
## Cases...Cumulative.20 Cases...Cumulative.31 Cases...Cumulative.22 Cases...Cumulative.23 Cases...Cumulative.24 Cases...Cumulative.25 Cases...Cumulative.28 Cases...Cumulative.29
## 1 66 116 70 71 77 82 93 99
## 2 11 18 13 13 13 13 17 17
## 3 5 9 6 6 6 6 6 8
## 4 17 25 22 23 24 24 25 25
## 5 155 291 187 194 198 215 247 263
## 6 20 46 22 27 27 27 36 39
## Cases...Cumulative.30 Cases...Cumulative.32 Cases...Cumulative.33 Cases...Cumulative.26 Cases...Cumulative.27 Cases...Cumulative.36 Cases...Cumulative.38 Cases...Cumulative.21
## 1 107 120 128 86 89 162 205 68
## 2 18 18 19 14 15 20 21 13
## 3 9 10 11 6 6 11 13 6
## 4 25 25 25 25 25 31 31 20
## 5 277 304 312 229 235 354 379 169
## 6 46 49 55 29 33 63 67 20
## Cases...Cumulative.35 Cases...Cumulative.37 Cases...Cumulative.40 geometry TotHousing
## 1 148 197 217 MULTIPOLYGON (((1177741 190... 1
## 2 20 20 22 MULTIPOLYGON (((1181225 190... 0
## 3 11 13 14 MULTIPOLYGON (((1179498 190... 0
## 4 30 31 31 MULTIPOLYGON (((1174762 189... 0
## 5 333 364 399 MULTIPOLYGON (((1178340 189... 1
## 6 61 64 76 MULTIPOLYGON (((1174680 190... 0A.7 Inspect Data
A.7.1 Thematic Maps
To inspect data from a spatial perspective, we can create a series of choropleth maps.
Example 1. Number of Affordable Housing Developments per Zip Code
Choose the variable “TotHousing” to map total developments per zip code, as we calculated previously. Here we’ll map using Jenks data breaks, with a Blue to Purple palette, and four bins (or groups of data to be plotted). A histogram of the data is plotted, visualizing where breaks occured in the data to generate the map.
tmap_mode("plot")
tm_shape(Chi_Zipsf.3435) + tm_polygons("TotHousing", legend.hist = TRUE, style="jenks", pal="BuPu", n=4, title = "Housing Dev.") +
tm_layout(legend.outside = TRUE, legend.outside.position = "right")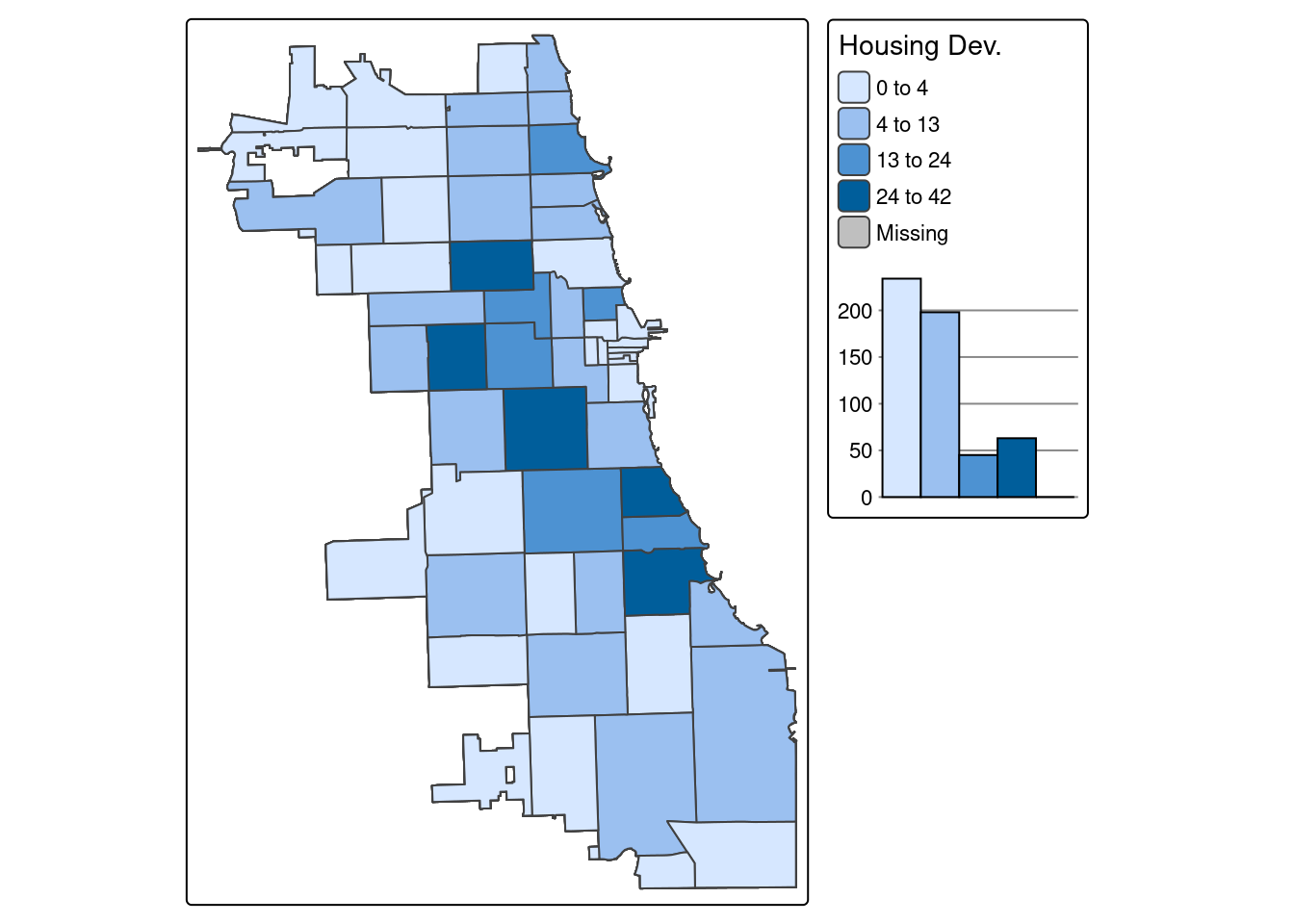
Note: You may get an error about missing the package ggplot2. To address this, just install that package and then rerun the command above.
Example 2. Number of COVID-19 Cases per Zip Code
Let’s do the same, but plut a different variable. Select a different variable name as your parameter in the ‘tm_fill’ parameter.
tm_shape(Chi_Zipsf.3435) + tm_polygons("Case.Rate...Cumulative",
legend.hist = TRUE, style="jenks",
pal="BuPu", n=4, title = "COVID Case Rate") +
tm_layout(legend.outside = TRUE, legend.outside.position = "right")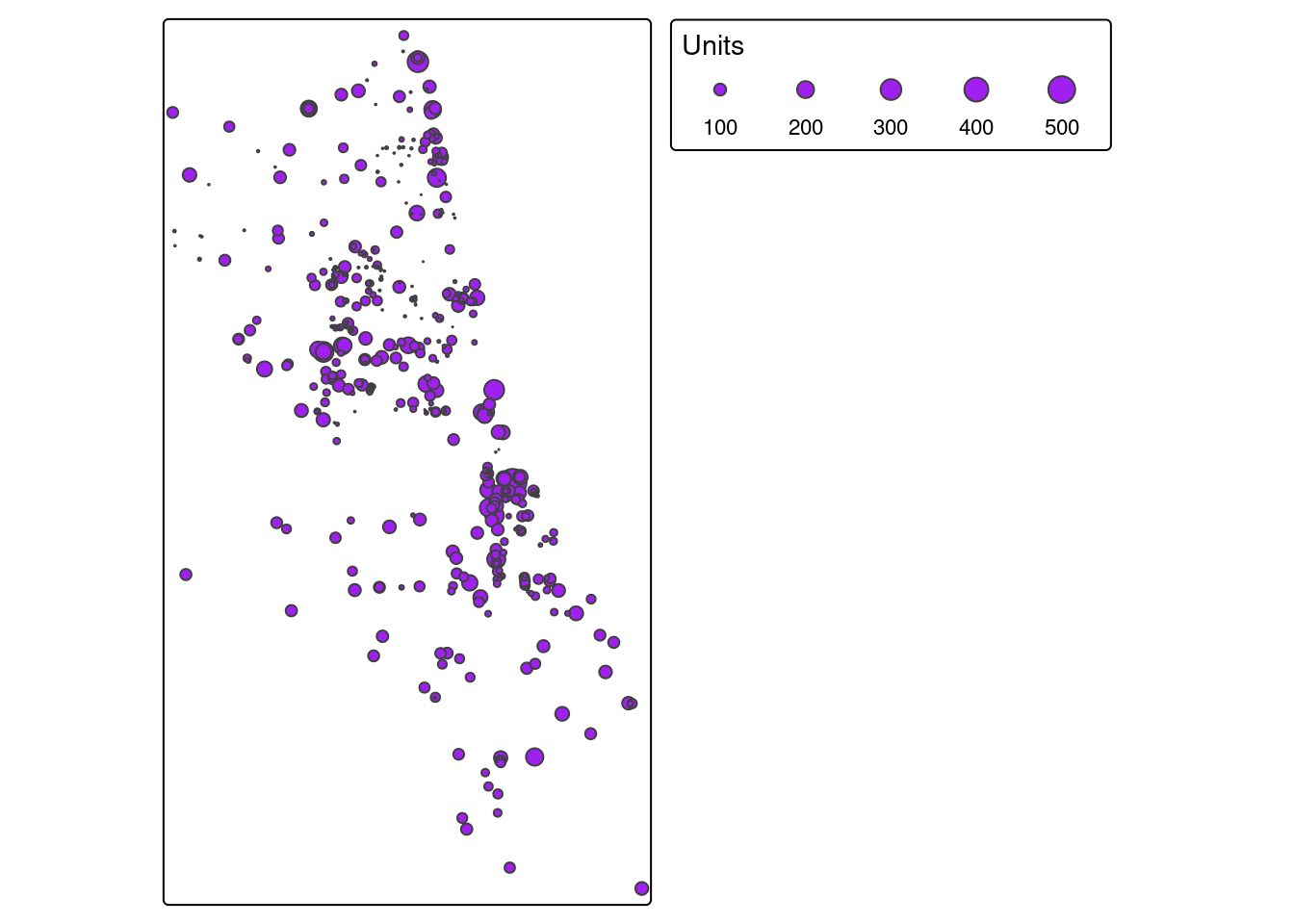
A.7.2 Map Overlay
Example 1. Affordable Housing Developments & Zip Code Boundaries
We previously translated the housing dataset from a CSV to a spatial object. Let’s take an attribute connect with each point, the total number of units per housing development, and visualize as a graduated symbology. Points with more units will be bigger, and not all places are weighted the same visually.
We use the “style” parameter to add a standard deviation data classification break. Check out tmap documentation for more options, like quantiles, natural jenks, or other options.
## ## ── tmap v3 code detected ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────## [v3->v4] `tm_bubbles()`: instead of `style = "sd"`, use fill.scale = `tm_scale_intervals()`.
## ℹ Migrate the argument(s) 'style' to 'tm_scale_intervals(<HERE>)'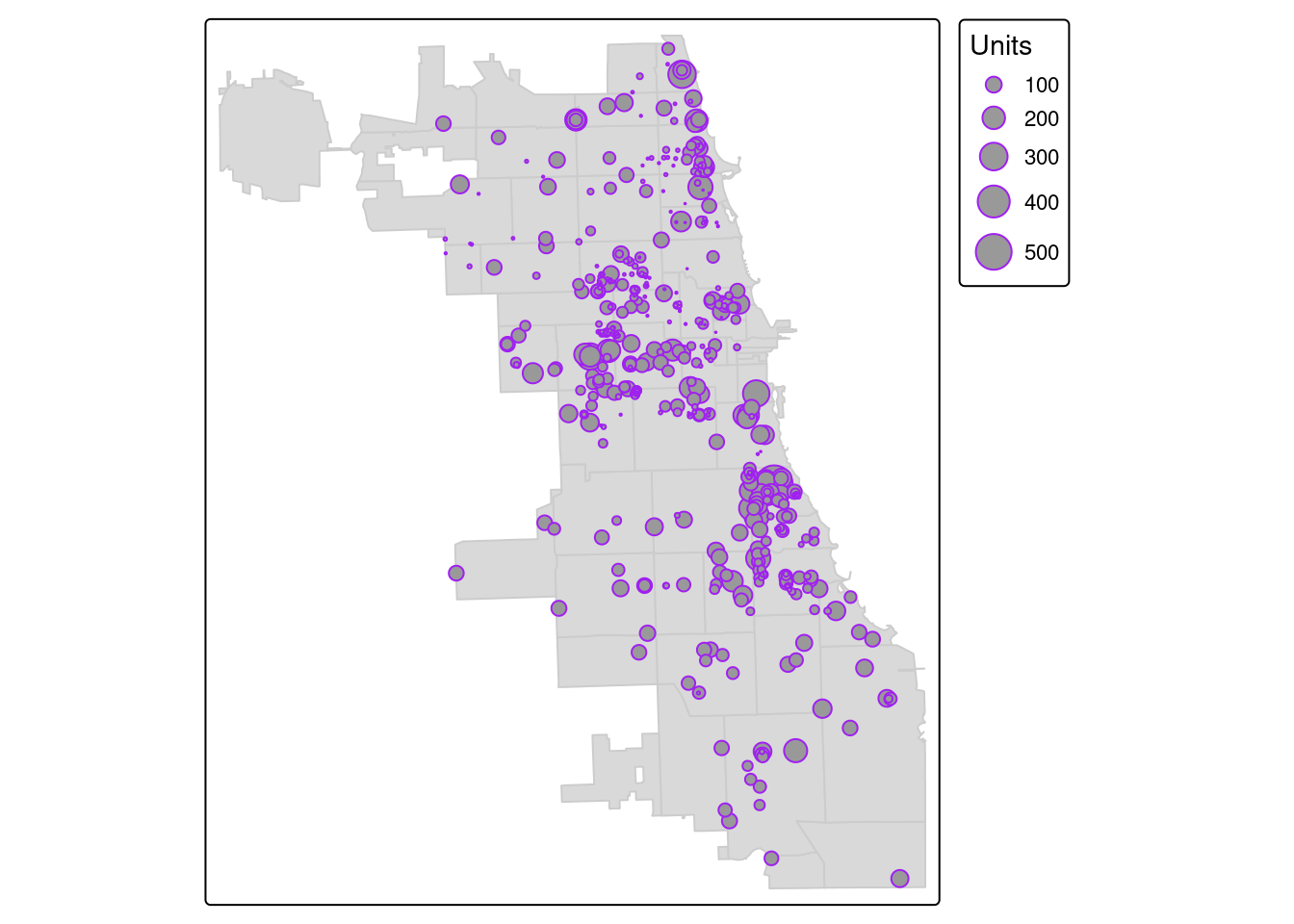
Then, let’s overlay that layer to the zip code boundary.
tm_shape(Chi_Zipsf.3435) + tm_polygons(col = "gray80") +
tm_shape(housing.3435) + tm_bubbles("Units", col = "purple") 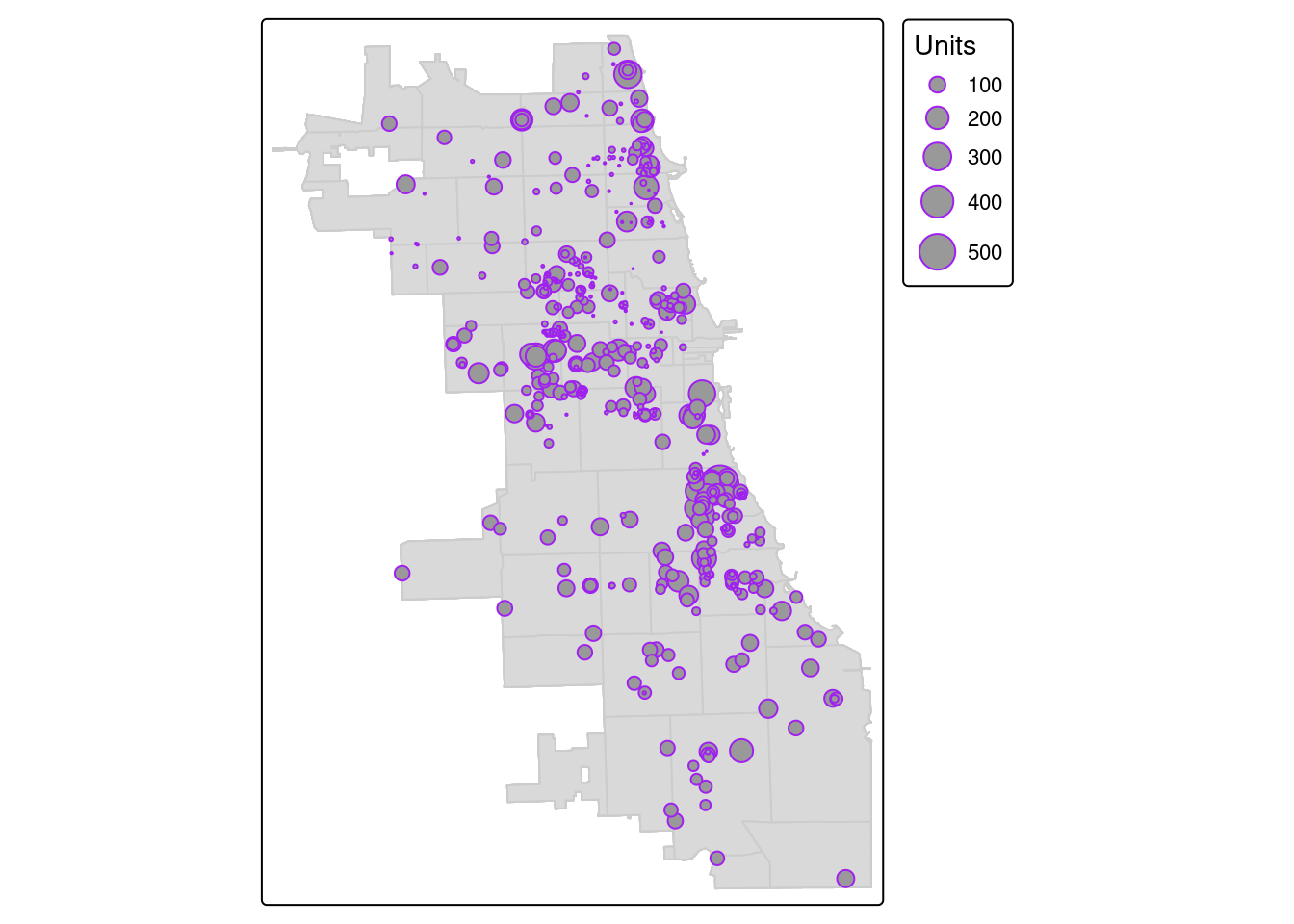
You can also color-code according to the total number of units. Here, we’ll add a palette using a “viridis” color scheme, as a graduate color point map. For extra style, we’ll add labels to each zip code, update with a new basemap, and make the whole map interactive.
## ℹ tmap mode set to "view".#Add Basemap
tm_basemap("Esri.WorldGrayCanvas") +
#Add First Layer, Style
tm_shape(Chi_Zipsf.3435) + tm_borders(col = "gray10") +
tm_text("zip", size = 0.7) +
#Add Second Layer, Style
tm_shape(housing.3435) +
tm_bubbles( col = "Units", style = "quantile",
pal = "viridis", size = 0.1) ##
## ── tmap v3 code detected ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
## [v3->v4] `tm_bubbles()`: instead of `style = "quantile"`, use fill.scale = `tm_scale_intervals()`.
## ℹ Migrate the argument(s) 'style' to 'tm_scale_intervals(<HERE>)'[tm_bubbles()] Argument `pal` unknown.